Cancer is a worldwide problem and it affects millions of people. Unfortunately there is still not a cure for cancer and this is due to the fact that each cancer affects different tissues of the body, which in turn are affected by different pathways within the body. Cancer is caused by a mutation within the DNA of a cell, which causes it to continue to replicate without stopping, and the cell receives the signal to do so through an epigenetic mechanism of action. This is accomplished by an attachment or detachment of a methyl group to histones that control the expression of genes along the DNA. There are certain methylation states of histones that control cellular replication. The methylation is used to maintain homeostasis for cellular replication, but if there is a mutation that occurs, or there is not proper nutrients being taken up in the diet, it could lead to a continuation of cellular growth leading to tumors and metastasis throughout the body. There are some drugs being tested that inhibit demethylation of histone receptors, and they have shown some promise for cancer treatment. Lunasin is a soybean protein that has also been shown to be a bioactive molecule that naturally controls the homeostasis of the methylation states of histones, specifically H3 and H4 histones. This is of novel importance because Lunasin can potentially be used as a natural cancer treatment or used as a natural method to prevent the development and spread of cancer.
Introduction
Epigenetics has become a very popular field of research within academic research in the last twenty years. It has been seen to play a very important role in hypomethylation, hypermethylation, chromatin modification, and loss of imprinting mechanisms on the genome. All of these genomic mechanisms are extremely important in understanding the mechanisms that are going on during the development of cancer (Feinberg, 2004). Each of these mechanisms are directly affected by the epigenome, which is the histones that affect the activity of the genome. The epigenome directly influences the expression of genes found within deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). This means that the epigenome can turn the expression of a gene “on” or “off”. Epigenetics is the study of the mechanisms of action that are not genotypical but rather phenotypical. This means that it does not alter the coding sequence but just the way that it is expressed. It studies the gene expression within the DNA sequence. The phenotypical changes occur depending on a gene being expressed or not. The gene expression is usually altered because of DNA methylation and histone modification (Feinberg, 2006).
The study of epigenetics is very attractive for potentially finding a better way to prevent and treat cancer. Cancer has its mechanism of action based within an epigenetic disruption of progenitor cells that are mediated by genes that encourage uncontrolled cellular division (Feinberg, 2006). This gene expression is controlled by the methylation of histone receptors upon DNA. If there were a way to directly control the methylation of the histone receptors it could likely lead to a better treatment and prevention of cancer. In fact it has been shown that transcriptional inactivation occurs due to the methylation occurring on the histone receptors. However, understanding the purpose of the different methylation states that control the expression of the genes is what is of the most concern, because it would enable us to understand what effect methylation or demethylation would have on the expression of a particular gene (Jones, 2003). The traditional understanding of cancer has always been that there is an accumulation of alterations that occur within oncogenes and tumor-suppressor genes that cause the uncontrolled tissue growth that will eventually lead to tumors. However, with the advent of epigenetic research it has been shown that there are epigenetic alterations that contribute to carcinogenesis and these alterations can be caused by different environmental and dietary exposures (Virani, 2012). If diet, exercise, and environment have an important role in epigenetics, that means that positive lifestyle changes in these areas would lead to a decrease in cancer development. It is beginning to move into the direction that epigenetic therapy or supplements will lead to a better clinical treatment and prevention of cancer. This review will discuss the epigenetic mechanisms of action that are affected by cancer, some of the synthetic epigenetic drugs that are currently being used to treat cancer, and natural cancer treatment and prevention in the form of a soy peptide called Lunasin.
Epigenetic Mechanisms of Action
Cancer is a genetic disease, more specifically it is a disease that is caused by specific genes that develop mutations and it then leads to uncontrolled cellular division. The pathways that are used during the process of mutation are of great concern because if they are better understood this may lead to the further development of cancer treatments that would better treat and prevent cancer (Vogelstein, 2004). In 1975 it was hypothesized that methylation was the mechanism that was used in order to allow a particular gene expression to remain stable during cellular division. Methylation of deoxyribonucleic acid (DNA) results in the chromatin (a complex of DNA, protein, and RNA) being placed into a stable state. The stable state of the chromatin means that DNA transcription is not happening. Methylation is achieved through protein components called histones that modify the behavior of nucleosomes (a unit of DNA packaging with around eight histone protein complexes which control gene expression on the DNA) to remain stable during cellular replication (Jaenisch, 2003). The tails of the histones project out from the nucleosome and serve as indicators within the cell to trigger a covalent posttranslational modification to occur. Some of the modifications are acetylation, methylation, phosphorylation, sumoylation, and ubiquitination. Each one of the modifications causes the chromatin to condense in a particular way depending upon the modification. The modification ultimately determines the histone code that regulates gene expression. So each one of these posttranslational modifications remodel the chromatin in a specific way that determines the epigenetic inheritance of the mechanisms for gene expression. The histone that is responsible for causing the effect on the chromatin has a particular place on the DNA strand. This means that each histone has the ability to cause a modification on the chromatin that causes a specific gene to be inhibited or expressed depending upon the location of the histone on the DNA (Thiagalingam, 2003).
Histone deacetylases (HDACs) are the enzymes that are responsible for removing the acetyl groups that are attached to the histones. It has been shown that histone acetylation weakens the influence that the histone has on gene expression. Histone acetylation causes a loss of stability within the chromatin of the cell causing an increase in transcriptional activity. Histone deacetylases cause the repression of transcription (Struhl, 1998). Since the histones are directly responsible for affecting gene expression, and they are directly affected by the histone deacetylases, there are many drugs that are in the development stages that target them specifically for cancer prevention. Histone deacetylase inhibitors (HDACi) are one of the drugs that appears to be showing the greatest amount of promise (Minucci, 2006). There are three different classes of HDACs that have been found in humans that are based upon their homology to yeast histone deacetylases, but the third class is the only one that has been found to be related to cancer (Jaenisch, 2003).
The third class of HDACs are shown to cause or affect chromatin silencing, cellular metabolism, and aging. Most of the class III HDACs are around 300–400 amino acids in length. This class of HDACs is still being heavily explored and there is not that much known about their mechanisms of action. However, class III HDAC has been shown to be associated with a repression of the expression of a particular gene that causes apoptosis when DNA is damaged (Thiagalingam, 2003). This particular HDAC causes a transcriptional activation that prevents apoptosis. This makes class III HDACs a matter of significance when exploring cancer therapies because studying this particular mechanism of action may lead to a better treatment or therapy for cancer patients (Thiagalingam, 2003). More research is needed on possibly using class III HDAC’s as a cancer treatment, but they do show promise in promoting the expression of the gene responsible for activating apoptosis within a cell that has a DNA mutation like cancer.
There are other factors that affect the histone besides HDACs, and they can also trigger methylation. It has been shown that the methylation of histone H3 at Lys9 has been linked to gene silencing because it stabilizes the state of the chromatin as well. This basically allows for the cell to have a memory from which it is able to differentiate between the parts of the DNA that are methylated or not. Evidence has also been found that indicates that different states of chromatin are mutually reinforcing or mutually inhibiting cellular processes. The most common cellular process that is affected by the state of chromatin is transcription. The different states of chromatin either increase transcriptional activity or silence it (Jaenisch, 2003).
In cancer, it was found that there were low levels of DNA methylation present in tumors compared to the DNA methylation levels found in normal tissue (Esteller, 2008). There are three proposed mechanisms that explain the role of DNA hypomethylation for the development of a normal cell into a cancer cell. These are: generation of chromosomal instability, reactivation of transposable elements, and loss of imprinting (Esteller, 2008). In certain parts of the body, DNA methylation causes different mechanisms to occur. For example, hypomethylation suppresses the later stages of intestinal tumorigenesis, but promotes early precancerous lesions in the colon and liver through genomic deletions (Yamada, 2005). The difference in the DNA methylation states in cancer is different depending on the location of the tissue within the body (Figure 1, Esteller, 2008).
Epigenetic Alterations in Tumor Progression
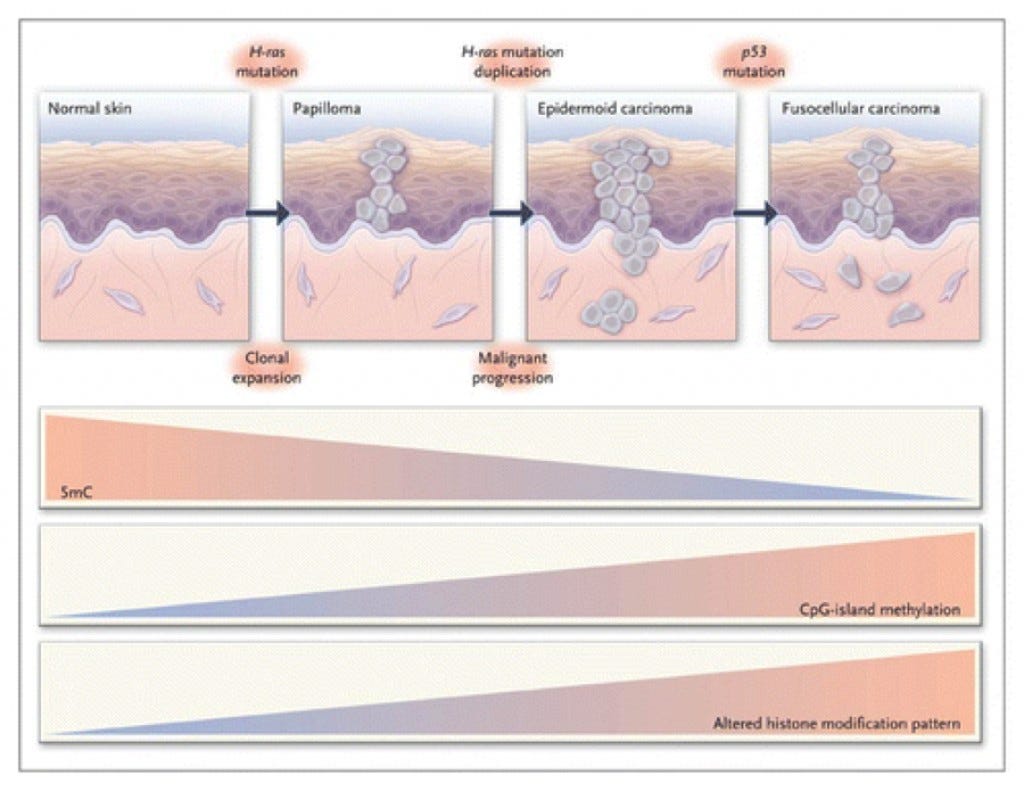
A multistage model of carcinogenesis in skin is shown. In conjunction with phenotypic cellular changes and the accumulation of genetic defects, there is a progressive loss of total DNA methylation content, an increased frequency of hypermethylated CpG islands, and an increased histone-modification imbalance in the development of the disease. H-ras denots Harvey-ras oncogene, and 5mC 5-methyl-cytosine (Esteller, 2008).
In figure 1, it is seen that different tissues within the body display different states and degrees of DNA hypomethylation. This is significant because it means that specific tissues will have different regulatory epigenetic mechanisms that occur under normal circumstances. If there is a mutation within DNA or significant epigenetic changes, it could lead to uncontrolled cellular growth eventually leading to cancer. However, in most cases when DNA is methylated it has been shown to cause the condensation of chromatin which further prevents transcription from occurring (Risch, 2008).
Mechanisms of Methylation
There are multiple genetic locations that have been shown to contribute to the development of cancer. Two of the genes that have been shown to be abnormally expressed in cancer are growth-promoting oncogenes and growth-suppressing tumor suppressor genes (Risch, 2008). The expression of these genes are amplified in cancer and are used as an indicator to show the location of the tumor suppressor genes found within DNA. In fact, a research study found that certain genetic lesions will cause epigenetic markers that are used to identify particular cancers, and in one specific case, lung cancer in particular (Risch, 2008). A very close relationship between genetic inheritance and environmental factors exists. This relationship is becoming increasingly more important as research continues into this area.
In human cancer DNA methylation of histones directly silences tumor suppressor genes. When methylation occurs it causes the chromatin to condense which does not allow for transcription to occur. The methylation is caused by protein complexes that contain methyl CpG-binding proteins (on the 5’ carbon where cytosine is next to guanine in sequence) and HDACs. When promoter sequences of DNA are methylated, it induces the deacetylation by HDACs, and this causes the chromatin to repress transcription. This suggests that methylation could be used as an identifier to determine whether or not genetic expressions have molecular abnormalities that could potentially lead to cancer. It has been found that out of the average number of 29,000 CpG-binding proteins found in human nucleosomes that 1,400 of them are methylated in patients with lung cancer (Risch, 2008). This would suggest that methylation of histones located on nucleosomes is directly related to the development of cancer.
The human genome can be affected by a number of epigenetic changes. Cytosine methyltransferases(DNMT) add a methyl group to each of the nucleosomes at CpG. These types of sequences are usually found in the promoter region, or the first intron of genes that are responsible for the upkeep of the cell as well as genes that are tissue specific. The sequence positions outlined above are almost always unmethylated in normal cells, but they could become methylated during development, differentiation, or cancer. Acetylation of histones on DNA is achieved by histone acetylases(HATs), and they are removed by HDACs. This basically means that HATs cause an acetyl group to attach to the histones and the HDACs are used to remove the attached acetyl groups when they receive the signal to do so. In order to learn how to better control the mechanisms of the activation and inhibition of HATs and HDACs, one particular experiment showed that HDACs are responsible for removing acetyl groups from the histones, and if they are inhibited by HDAC inhibitors this will lead to a growing number of acetyl groups that are attached to histones on the DNA (Thiagalingam, 2003). When there is an accumulation of acetyl groups on histones it leads to a decrease in the expression of genes that control cellular arrest, differentiation, and apoptosis. This is why it is believed that HDACis hold the most promise for looking for a better treatment for the treatment and prevention of cancer (Thiagalingam, 2003).
Epigenetic Changes in Cancer
There are three different ways that epigenetics play a role in cancer. They are DNA methylation, ribonucleic acid (RNA) — associated silencing, and histone modifications (Egger, 2004). Histone modifications have already been established as epigenetic modifiers, and the epigenetic changes occur by acetylation and methylation of lysine residues on the tail of the histones. Because lysine methylation can occur in situations that are monomeric, dimeric, or trimeric, and there are other posttranslational modifications that occur like acetylation, methylation, and phosphorylation, there is a lot of variation that can occur with molecular modification of histones. That is why it is being referred to as a ‘histone code’ by many researchers (Egger, 2004). If we understand what molecular modifications are occurring with the histones, then we can better understand how it could be changed to create a different outcome. In essence we could control the cellular replication and behavior by controlling its epigenetic processes.
DNA methylation is promising because it can be passed on to the next generation of cells. This means that the methylation states of different histones will be the same in the following generations unless an epigenetic modification occurs (Baylin, 1997). It also has the potential of being used as an identifier to show which nucleosomes are affected for the particular type of cancer. There are tumor specific CpG sites where hypermethylation occurs, and particular cancers also have a larger or smaller number of CpG sites that are methylated. However, it is still unclear why some CpG sites are methylated and others are not (Esteller, 2007). It is evident that methylation is a very important part of DNA expression, but it is still unclear how many CpG sites are not methylated. The methylation serves as a marker but unfortunately it does not tell the number histones that are left unmethylated. However, one potential way to solve this problem may be to study the prevalence of MicroRNAs. They are short non-coding RNA’s that interact with specific mRNAs and they can induce mRNA degradation or translational inhibition. MicroRNAs can also serve as oncogenes or tumor-suppressor genes depending the epigenetic modifications that have occurred for a particular gene (Lujambio, 2008). It has been found that a genetic lesion that occurs in cancer cells can create an epigenetic alteration in the human genome. This is why it is important to continue to do research on HDAC inhibitors and histone methyltransferase (HMT) inhibitors. HMT inhibitors have been showing the most promise because they only have two possible situations, the histone is either methylated or unmethylated (Lyko, 2005). For HDACs it is not as straight-forward because there are other forms of histone modifications that can occur at different parts of the nucleosomes, and it can also be affected by acetylation, methylation, and phosphorylation.
The methylation of the genome causes three different mechanisms to occur. These mechanisms are chromosomal instability, reactivation of transposable elements, and loss of imprinting (Esteller, 2002). Each of these together causes a loss of heterozygosity which in turn causes an increase in tumor metastasis all throughout the body. This is important in the study of cancer because it has established that methylation has been linked to increasing tumor metastasis (Esteller, 2002). HATs are activators of transcription and they rearrange the promoter region of the chromatin to cause the expression of a gene. HDACs deacetylate the histone tails and cause a repression of transcription activity. Both of these enzymes are needed in order to regulate the epigenetic activity of the genome (Esteller, 2002).
A particular study has shown that a high number of HDACs may be an early step in the process towards developing cancer (Virani, 2012). HDAC inhibitors are one of the most promising forms of cancer treatment. Currently there are two HDAC inhibitors that are approved by the FDA for the treatment of cutaneous T-cell lymphoma. The first is suberoylanilide hydroxamic acid (virinostat) and the second is romidepsin (Virani, 2012). One of the reasons that epigenetics is so promising for research is that epigenetic changes are reversible where genetic changes are not. If epigenetic changes can be induced by diet and exercise that positively affect the genome, it would be especially helpful towards cancer patients. There are some scientists working on that very idea.
Natural Cancer Prevention and Treatment
It has been shown by epidemiological studies that the incidence of cancer varies greatly depending upon the geographical location (Meisel, 2004). It was further discovered through research that the geographical areas where the incidence in an area was lower, that people had a diet that was heavily centered on plant foods (Meisel, 2004). This could suggest that diet plays a role in the development of cancer. Some peptides found in foods act to reduce oxidative stress within the body, and some of these oxidative stresses are related to cancer (Meisel, 2004). Some peptides stimulate the immune system that further activates cytomodulatory mechanisms that prevent cancer cell growth. Some of the cytomodulatory mechanisms that are triggered are the production of antimicrobial peptides that are produced to kill sensitive microorganisms, the production of angiotensin-1-converting enzyme inhibitory peptides, and the production of opioid peptides (opioid receptor ligands that modulate the absorption processes in the intestinal tract) (Meisel, 2004).
There are certain foods that have bioactive components that protect the body from carcinogens. One such food is soybean, and it protects the body because it contains phytochemicals that promote antioxidant activity, and it also inhibits proteases that cause the initiation and promotion of cancer. It inhibits proteases by the Bowman-Birk protease inhibitor (BBI) which is a component of the soybeans. There are other soybean proteins that show promise in preventing cancer, but one in particular and that would be a protein named lunasin (Hernandez, 2009). It has anti-inflammatory and antioxidant effects that possibly contribute to a chemoprotective action against cancer (Cam, 2013). It was found that lunasin elicits a reduction in intracellular Ca2+in LPS-induced macrophages, it also causes the internalization of lunasin to increase during times of inflammation, and it may also be responsible for the attenuation of cardiovascular disease risk factors by interacting with pathways that are involved in endocytosis and inflammation (Cam, 2013). When ingested, lunasin has also been found in plasma, and this suggests that it reaches many different tissues where it can affect many physiological activities (Dia, 2009).
Lunasin was discovered during an experiment that was trying to enhance the quality of the proteins found within soybeans (Galvez, 1997). When lunasin was isolated and placed into mammalian cells it was found to disrupt mitosis, cause chromosomal fragmentation, and apoptosis. Lunasin is composed of 43 amino acids and it has an interesting molecular structure because there are 8 aspartic acid (Asp) residues at the carboxyl end of the protein. These Asp residues are responsible for the mitotic effects because this tail of 8 Asp residues bind to the histones and it affects the chromatin in such a way that the kinetochore is unable to form and the microtubules do not attach to the centromeres. This eventually causes the cell to die because it is unable to replicate. It was further found that in the absence of carcinogens, lunasin does not appear to affect the cell morphology or proliferation, but just prevents the transformation of cells when there are carcinogens present (Hernandez, 2009).
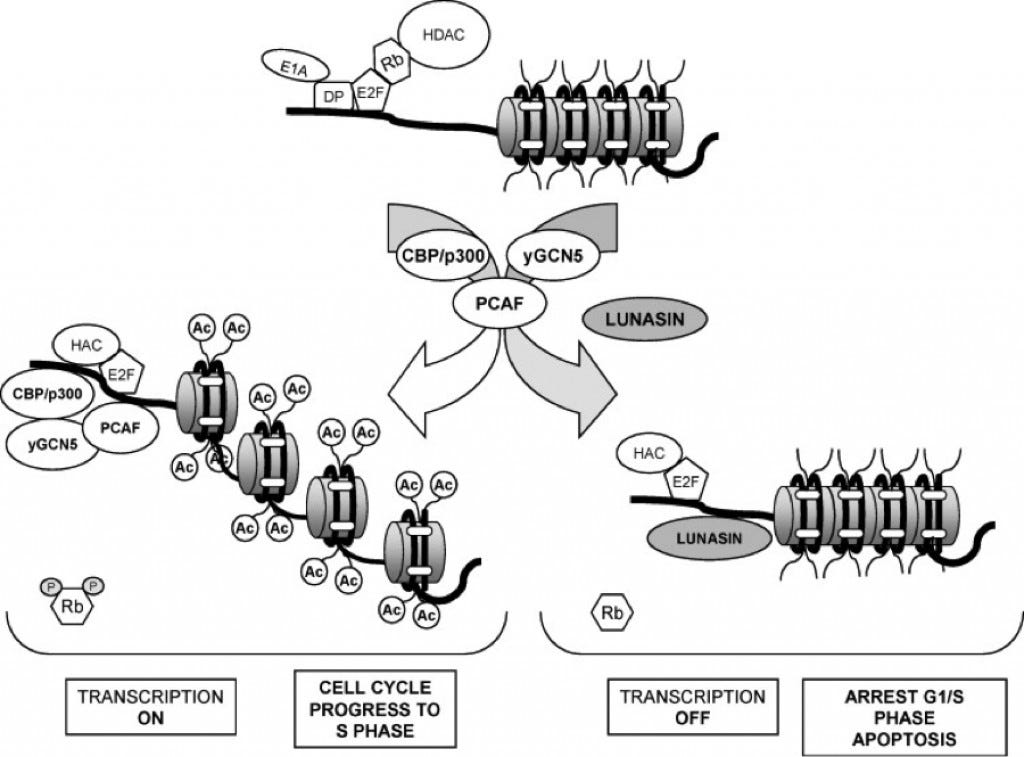
Lunasin has also been found to disrupt carcinogenesis and oncogenesis by common mechanisms that change the chromatin status (Hernandez, 2009). They are histone acetylation and deacetylation by HATs and HDACs. These two processes are heavily involved in controlling chromatin behavior and they also play an important role as tumor suppressors. There are two histones, H3 and H4, that are referred to as the core histones because they have been found to be the most important in cancer research (Hernandez, 2009). In normal conditions within the cell, H3 and H4 histones are usually deacetylated. During an experiment where cells were treated with a known HDAC inhibitor, sodium butyrate, histone acetylation did occur. When the cells were treated with sodium butyrate and lunasin collectively it was found that acetylation was inhibited (Hernandez, 2009). This suggests that lunasin plays an important role in preventing acetylation of the histones. In fact, lunasin operates as a protein that inhibits histone acetylation and that means it also operates as a natural tumor suppressor. Lunasin competes with HATs in binding to the deacetylated core histones, and if it attaches to H3 and H4 it represses cell cycle and could even lead to apoptosis of the cell (Figure 1)(Hernandez, 2009). This is very important if the cell being targeted was a cancer cell because the cell would die before it could become a threat to the rest of the body.
Effect of Lunasin on Histones
In another experiment, lunasin was topically applied to the skin of rodents with tumors, and it was shown to reduce the size of the tumor and prevent further metastasis and growth (Galvez, 2001). Lunasin is a novel soybean protein and it does seem to have promise for being a chemopreventive agent against cancer.
Cancer is heavily related to abnormal genetic and epigenetic changes that occur within the cells of the body. Lunasin is a component of soybeans and it has been shown to have cancer preventing properties due to its ability to act as a natural HDAC inhibitor (Galvez, 2001). The acetylation causes an increase in DNA transcription, and if it is inhibited it will cause a repression of the transcription. Lunasin acts as a natural HDAC inhibitor by inhibiting HDAC’s from removing the acetyl group. There are other bioactive foods that affect DNA methylation but none that are as promising as lunasin. In fact, lunasin has been shown to enhance the reduction of tumors in lymphoma when immunization occurs with Ovalbumin (Tung, 2014). In mice that were immunized with lunasin and Ovalbumin it was shown to have greater tumor inhibition than without the lunasin (Figure 2., Tung 2014). Tumor volume was seen to be drastically decreased when lunasin was used as an adjuvant compared to the phosphate buffered saline (PBS)(Figure 2, panel A). It was also determined that the higher concentrations of lunasin was able to be absorbed into the nucleus of the cells in the rodent and that it caused a greater response of the CD8-specific T-Lymphocytes (Figure 2, panel B and C). This is a novel discovery because the results from this experiment show that lunasin is the first natural adjuvant that has been used to cause an immune response compared to using a toxin in order to create an immune response (Tung, 2014).
Effects of Lunasin With Ovalbumin as an Adjuvant in Immunization
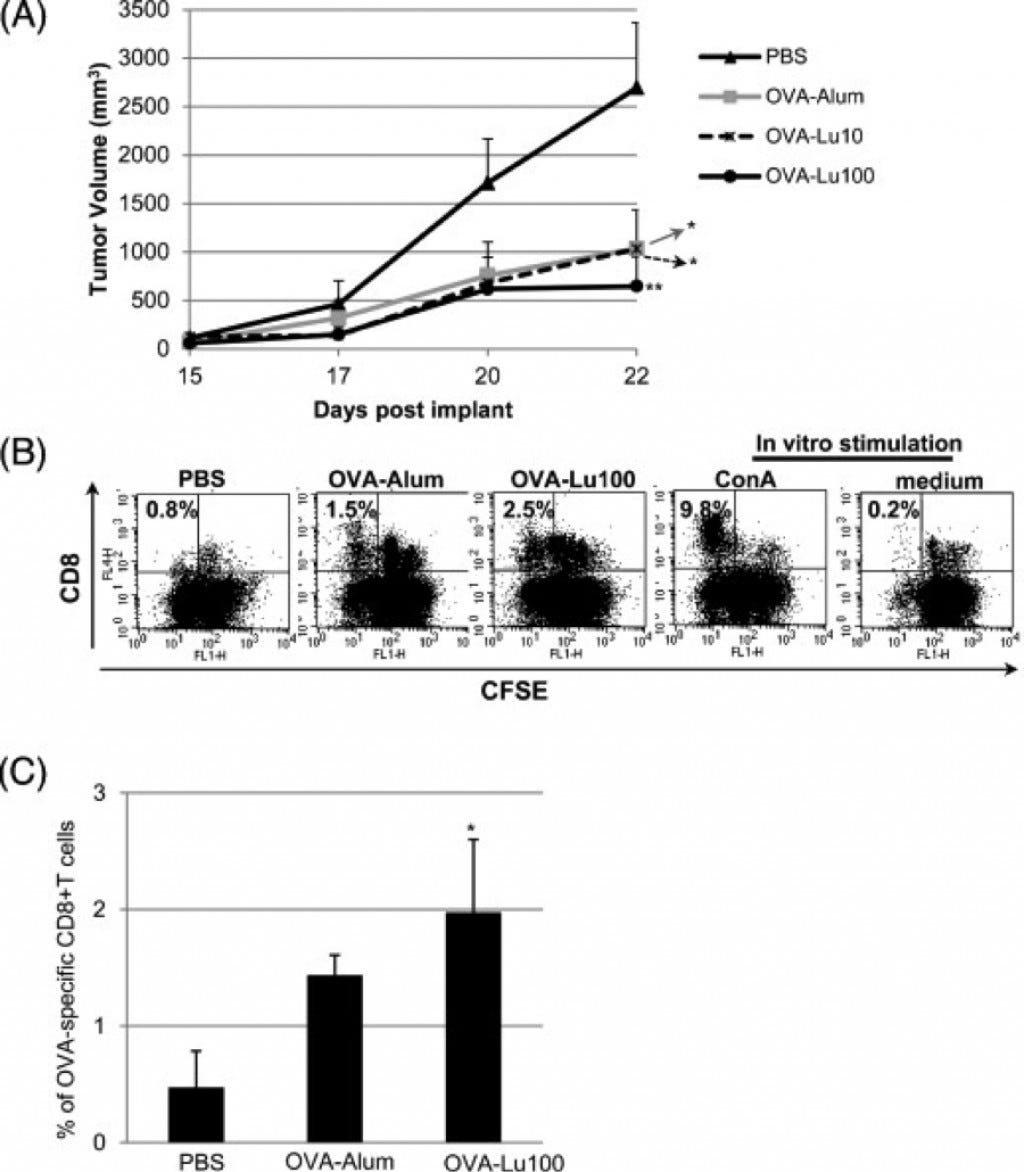
(A) Tumor volumes were measured from day 15 after injection through day 22. Data represent the mean ± SEM from 5 mice per group. A mixed model with repeated measure to the data was developed using PROC MIXED in SAS program followed by pairwise comparison test of the mean differences among treatments by different days. *P ≤ 0.05; **P ≤ 0.01, relative to PBS-vaccinated group. Results shown are representative from 2 independent experiments with similar profiles. (B) All mice were euthanized 22 days following tumor injection. Spleens collected from these mice were processed into single-cell suspensions followed by labeling with carboxyfluorescein diacetate succinimidyl ester (CFSE). CFSE-labeled splenocytes were then stimulated with OVA (100 μg/ml) to induce proliferation of OVA-specific T cells. For the in vitro controls, CSFE-labeled splenocytes were cultured in medium only as a negative control, or treated with Concanavalin A (Con A) (2.5 μg/ml) for polyclonal proliferation as a positive control. Proliferation of CD8+ T cells was determined from diluted CFSE using flow cytometry. A representative dot plot shows the percentage of diluted CFSE at the upper left quadrant. (C) The averaged percentage of proliferation from OVA-specific CD8+ T cells is shown as mean ± SD from 3 mice per group. *P ≤ 0.05, relative to PBS-vaccinated group (Tung, 2014).
The methyl groups that are used in the epigenetic mechanisms for methylation are likely acquired in our diets or are made as secondary components from our dietary intake (Ross, 2003). In fact, dietary factors directly influence the supply of methyl groups because they influence the formation of S-Adenosyl methionine (SAM) which is a cosubstrate that is used for methyl group transfers in the body. Dietary factors also can have an impact on DNA methyltransferase and demethylation activity (Ross, 2003). For example, some essential components of a healthy diet are folate, vitamin B12, vitamin B6, vitamin B2, methionine, and choline. Each of these influence SAM in one form or another and by influencing SAM it also influences DNA methylation. This could cause a number of different cellular affects to occur due to the epigenetic changes that would occur with the methylation or demethylation. Diet affects apoptosis, cell cycle, differentiation, inflammation, angiogenesis, DNA repair, and carcinogen metabolism (Ross, 2003). Each of these are also affected by methylation patterns of histones which is one of the reasons why diet plays such a major role in the body. Without a healthy diet the body will be unable to carry out DNA methylation or demethylation properly because the cosubstrate SAM will not adequately form (Ross, 2003). More research is still needed in this area, but it is becoming more evident that with a healthy diet high in bioactive foods that there is less likelihood of developing cancer. Also, if an individual has cancer it will be of benefit to them to have a diet that is high in bioactive foods to reduce the negative epigenetic changes from occurring.
Conclusion
Cancer can become a debilitating and sometimes fatal disease. Unfortunately there are still no widely accepted natural therapies or treatments that are able to successfully treat and prevent cancer. However, scientists from all across the world are working towards the common goal of finding better ways to treat and prevent it. Cancer is a very complicated disease because it has the potential to affect so many different areas of the human body. In some cases, there is not just one area in the body that cancer is destroying, but rather an alteration of the chromatin can be occurring in every affected cell. If the affected cells continue to replicate with the mutations within the chromatin, unproliferated cellular growth begins to occur at the site of the cellular mutation. As the disease progresses metastasis would occur because the affected cells travel through the blood stream to the other parts of the body and affect new areas of healthy tissue. If nothing is done to prevent the mutations from occurring and nothing is done to prevent metastasis, an individual with cancer will usually die within a short period of time.
It has been shown how changes in the histones can modify the chromatin in a particular cell to affect the expression of a specific gene (Baylin, 1997). Gene expression is heavily dependent upon the epigenetic mechanisms that regulate the activity of the cell (Thiagalingam, 2003). A cell responds differently to methylation of histones when compared to demethylation, and it also responds differently to acetylation/deacetylation and phosphorylation/dephosphorylation (Jaenisch, 2003). The body does this to try to maintain homeostasis throughout every one of its systems. The “brain” of the cell is known as the DNA and it is located within the nucleus. When there is something out of balance or a need within the body a signal is created that causes certain epigenetic processes to occur by affecting the histones wrapped around the DNA (Minucci, 2006). Diet also plays a major role in helping to produce components that are used within the epigenetic signaling pathway (Vogelstein, 2004). Without a healthy diet the proper components for cellular regulation will not be able to be made like SAM, and there may be an increase in hypermethylation or hypomethylation that causes unproliferated cellular growth to occur (Ross, 2003). Lunasin is promising as a natural cancer therapy and treatment that could potentially be used to help patients around the world (Galvez, 2001). The research on its potential uses and its effects within the human body are ongoing. It could potentially revolutionize the way that cancer is treated, but it can also help treat and prevent other diseases that occur because of a dysregulation of the epigenome.
References
Baylin, S. B.. “DNA METHYLATION: Tying It All Together: Epigenetics, Genetics, Cell Cycle, and Cancer.” Science 277.5334 (1997): 1948–1949. Print.
Cam, Anthony, Mayandi Sivaguru, Elvira Gonzalez De Mejia, and Christian Schulz. “Endocytic Mechanism of Internalization of Dietary Peptide Lunasin into Macrophages in Inflammatory Condition Associated with Cardiovascular Disease.” PLoS ONE 8.9 (2013): e72115. Print.
Dia, Vermont P., Sofia Torres, Ben O. De Lumen, John W. Erdman, and Elvira Gonzalez De Mejia. “Presence of Lunasin in Plasma of Men after Soy Protein Consumption.” Journal of Agricultural and Food Chemistry 57.4 (2009): 1260–1266. Print.
Egger, Gerda, Gangning Liang, Ana Aparicio, and Peter A. Jones. “Epigenetics In Human Disease And Prospects For Epigenetic Therapy.” Nature 429.6990 (2004): 457–463. Print.
Esteller, M.. “Epigenetic gene silencing in cancer: the DNA hypermethylome.” Human Molecular Genetics 16.R1 (2007): R50-R59. Print.
Esteller, M. (2008). “Epigenetics In Cancer.” New England Journal of Medicine, 1148–1159. Print
Feinberg, Andrew P., and Benjamin Tycko. “The history of cancer epigenetics.” Nature Reviews Cancer 4.2 (2004): 143–153. Print.
Feinberg, Andrew P., Rolf Ohlsson, and Steven Henikoff. “The epigenetic progenitor origin of human cancer.” Nature Reviews Genetics 7.1 (2006): 21–33. Print.
Galvez A.F., N. Chen, J. Macasieb, B.O. de Lumen. “Chemopreventive property of a soybean peptide (Lunasin) that binds to deacetylated histones and inhibit acetylation.” Cancer Res, 61 (2001), pp. 7473–7478
Galvez AF, Revilleza MJR, de Lumen BO. A novel methionine-rich protein from soybean cotyledon: cloning and characeterization of cDNA (accession No. AF005030). Plant Register #PGR97–103. Plant Physiol. 1997;114:1567–9. Print
Hernández-Ledesma, Blanca, Chia-Chien Hsieh, and Ben O. De Lumen. “Lunasin, a novel seed peptide for cancer prevention.” Peptides 30.2 (2009): 426–430. Print.
Jaenisch, Rudolf, and Adrian Bird. “Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals.” Nature Genetics 33.3s (2003): 245–254. Print.
Jones, Peter A.. “Epigenetics in Carcinogenesis and Cancer Prevention.” Annals of the New York Academy of Sciences 983.1 (2003): 213–219. Print.
Lujambio, A., C. M. Croce, S. Ropero, A. Villanueva, G. A. Calin, M. Esteller, S. A. Eccles, W. M. Gallagher, W. J. Faller, M. S. Nicoloso, S. Rossi, L. M. Montuenga, D. Blanco, and M. Sanchez-Cespedes. “A MicroRNA DNA Methylation Signature For Human Cancer Metastasis.” Proceedings of the National Academy of Sciences 105.36 (2008): 13556–13561. Print.
Lyko, F., and R. Brown. “DNA Methyltransferase Inhibitors and the Development of Epigenetic Cancer Therapies.” JNCI Journal of the National Cancer Institute 97.20 (2005): 1498–1506. Print.
Meisel, Hans. “Multifunctional peptides encrypted in milk proteins.” Biofactors 21.1–4 (2004): 55–61. Print.
Minucci, Saverio, and Pier Giuseppe Pelicci. “Histone Deacetylase Inhibitors And The Promise Of Epigenetic (and More) Treatments For Cancer.” Nature Reviews Cancer 6.1 (2006): 38–51. Print.
Risch, Angela, and Christoph Plass. “Lung cancer epigenetics and genetics.” International Journal of Cancer 123.1 (2008): 1–7. Print.
Rodríguez-Paredes, Manuel, and Manel Esteller. “Cancer epigenetics reaches mainstream oncology.” Nature Medicine unknown (2011): 330–339. Print.
Ross, Sharon A.. “Diet and DNA Methylation Interactions in Cancer Prevention.” Annals of the New York Academy of Sciences 983.1 (2003): 197–207. Print.
Struhl, K.. “Histone acetylation and transcriptional regulatory mechanisms.” GENES & DEVELOPMENT 12.5 (1998): 599–606. Print.
Thiagalingam, Sam, Kuang-Hung Cheng, Hyunjoo J. Lee, Nora Mineva, Arunthathi Thiagalingam, and Jose F. Ponte. “Histone Deacetylases: Unique Players in Shaping the Epigenetic Histone Code.” Annals of the New York Academy of Sciences 983.1 (2003): 84–100. Print.
Tung, Chun-Yu, Jie Sun, Baohua Zhou, Michael J. Robertson, Fang Li, Shuyu Yao, Morayo Jaja, Ling Han, David E. Lewis, and Hua-Chen Chang. “Activation of dendritic cell function by soypeptide lunasin as a novel vaccine adjuvant.” Vaccine 32.42 (2014): 5411–5419. Print.
Virani, S., J. A. Colacino, J. H. Kim, and L. S. Rozek. “Cancer Epigenetics: A Brief Review.” ILAR Journal 53.3–4 (2012): 359–369. Print.
Vogelstein, Bert, and Kenneth W Kinzler. “Cancer genes and the pathways they control.” Nature Medicine 10.8 (2004): 789–799. Print.
Widschwendter, Martin, Ian Jacobs, Peter W Laird, Joanne Young, Mihaela Campan, Daniel J Weisenberger, Christian Marth, Gilbert Spizzo, Elisabeth Mueller-Holzner, Daniel Egle, and Heidi Fiegl. “Epigenetic stem cell signature in cancer.” Nature Genetics 39.2 (2006): 157–158. Print.
Wolffe, A. P.. “Epigenetics: Regulation Through Repression.” Science 286.5439 (1999): 481–486. Print.
Yamada, Y. (2005). “Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis.” Proceedings of the National Academy of Sciences, 13580–13585. Print





